Articles
 WHAT'S SPECIAL ABOUT SPATIAL NETWORKS?
While a few proof-of-concept experiments have demonstrated the feasibility and potential of sequencing-based microscopy, there is still a great need for a principles-based understanding - how information can be preserved and communicated through molecular networks, and the theoretical and practical limitations governing data quality and resolution. Our work addresses this need by examining the deep connection between Euclidean geometry and spatially informative networks such as those produced in sequencing-based microscopy, but which also appear elsewhere such as high speed rail networks. We discovered that Euclidean geometric laws, including familiar formulas like the relationship between a sphere’s volume and its radius, have network-based counterparts that may be used to measure the consistency with which Euclidean geometry is approximated by any network. We have called this property “spatial coherence”, and show that it is a basic feature of spatial networks that can be measured without ground truth comparison or prior sample knowledge.
WHAT'S SPECIAL ABOUT SPATIAL NETWORKS?
While a few proof-of-concept experiments have demonstrated the feasibility and potential of sequencing-based microscopy, there is still a great need for a principles-based understanding - how information can be preserved and communicated through molecular networks, and the theoretical and practical limitations governing data quality and resolution. Our work addresses this need by examining the deep connection between Euclidean geometry and spatially informative networks such as those produced in sequencing-based microscopy, but which also appear elsewhere such as high speed rail networks. We discovered that Euclidean geometric laws, including familiar formulas like the relationship between a sphere’s volume and its radius, have network-based counterparts that may be used to measure the consistency with which Euclidean geometry is approximated by any network. We have called this property “spatial coherence”, and show that it is a basic feature of spatial networks that can be measured without ground truth comparison or prior sample knowledge.
D. F. Bonet, J. I. Blumenthal, S. Lang, S. K. Dahlberg, I. T. Hoffecker*; Spatial coherence of DNA barcode networks Patterns, 2025
 A SPATIAL NETWORK, HIDING IN PLAIN SIGHT
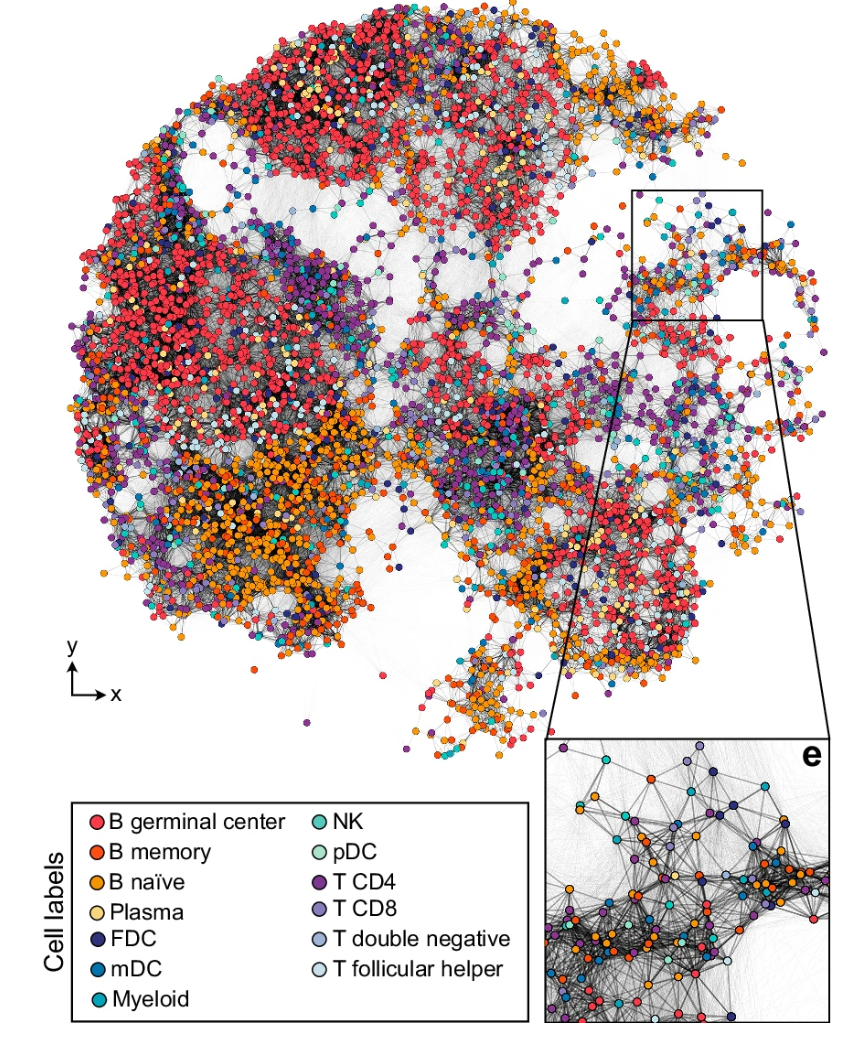
Together with Patrik Ståhl, an expert in spatial biology technologies, we made an exciting discovery when analyzing the publicly available data from Slide-tags, a spatial transcriptomics method recently developed by Chen and colleagues from the Broad Institute that is meant for coupling single nucleus sequencing to spatial maps. What we found was that the Slide-tags procedure, in which barcoded oligonucleotides are diffused into nearby tissue, leads to the formation of a spatially informative network of barcodes that essentially embeds spatial information into the structure of the sequencing data itself. The mix of barcodes recovered from each cell allows us to rebuild the network, whose structure reflects that of the spatial positions of the cells themselves. Or in other words, our team was able to computationally recover the relative spatial positions of cells entirely without the ground truth map that Chen and his team had obtained using a traditional microscopy-based decoding step. This spatially informative network information was hiding in plain sight in the Slide-tags data..
A SPATIAL NETWORK, HIDING IN PLAIN SIGHT
Together with Patrik Ståhl, an expert in spatial biology technologies, we made an exciting discovery when analyzing the publicly available data from Slide-tags, a spatial transcriptomics method recently developed by Chen and colleagues from the Broad Institute that is meant for coupling single nucleus sequencing to spatial maps. What we found was that the Slide-tags procedure, in which barcoded oligonucleotides are diffused into nearby tissue, leads to the formation of a spatially informative network of barcodes that essentially embeds spatial information into the structure of the sequencing data itself. The mix of barcodes recovered from each cell allows us to rebuild the network, whose structure reflects that of the spatial positions of the cells themselves. Or in other words, our team was able to computationally recover the relative spatial positions of cells entirely without the ground truth map that Chen and his team had obtained using a traditional microscopy-based decoding step. This spatially informative network information was hiding in plain sight in the Slide-tags data..
S. K. Dahlberg, D. F. Bonet, L. Franzén, P. S. Ståhl, I. T. Hoffecker*; Hidden network preserved in Slide-tags data allows reference-free spatial reconstruction
Nat. Comms., 2025
 MAPPING THE RNA SHAPE SWITCHES THAT CONTROL GENES
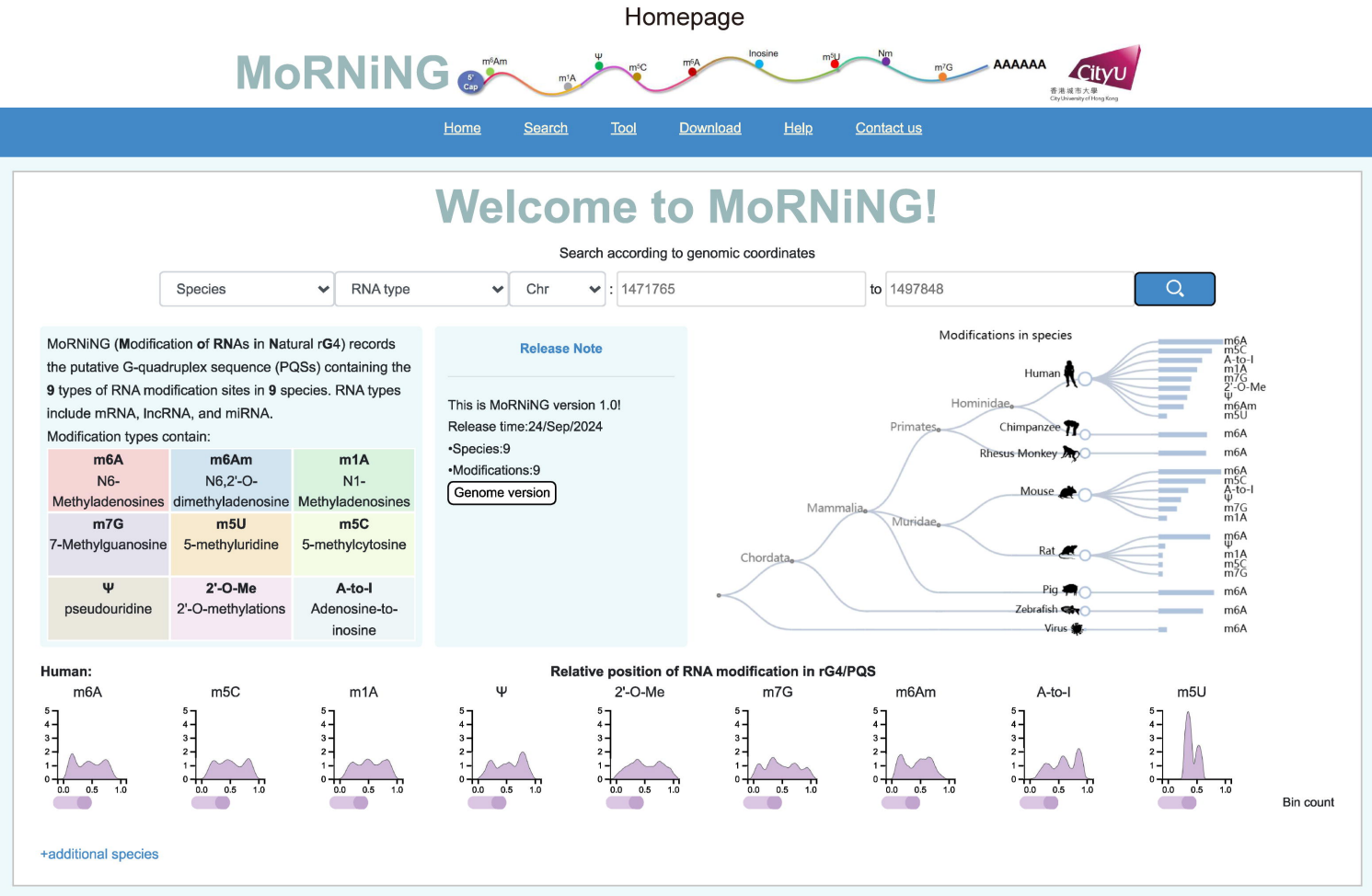
RNA isn't just a string of letters - it folds into tiny 3D shapes that help regulate almost every aspect of a cell's biology. The RNA G-quadruplex (rG4) is one of the more exotic forms that can arise - a knot-like structure when four guanines stack together. These shapes can act like molecular switches that influence how genes are read, how proteins bind, and how cells respond to signals. Interestingly, the same RNA segment can fold into different shapes depending on its chemical environment and the presence of chemical modifications.
In this work, led by our collaborator Jilin Zhang at City University of Hong Kong and colleagues, a new online database is established cataloging where RNA modifications fall within potential rG4 forming regions. Lab experiments showed that different modifications can shift the balance between RNA structures.
MAPPING THE RNA SHAPE SWITCHES THAT CONTROL GENES
RNA isn't just a string of letters - it folds into tiny 3D shapes that help regulate almost every aspect of a cell's biology. The RNA G-quadruplex (rG4) is one of the more exotic forms that can arise - a knot-like structure when four guanines stack together. These shapes can act like molecular switches that influence how genes are read, how proteins bind, and how cells respond to signals. Interestingly, the same RNA segment can fold into different shapes depending on its chemical environment and the presence of chemical modifications.
In this work, led by our collaborator Jilin Zhang at City University of Hong Kong and colleagues, a new online database is established cataloging where RNA modifications fall within potential rG4 forming regions. Lab experiments showed that different modifications can shift the balance between RNA structures.
Y. Zhou, S. Lyu, S.W. Liew, X. Mou, I.T. Hoffecker, J. Yan, Y. Li, C.K. Kwok, J. Zhang; Modification of RNAs in natural rG4 (MoRNiNG), A Database of RNA Modification Sites Associated with the Dynamics of RNA Secondary Structures BioRxiv, 2024
 DNA ORIGAMI STRUCTURAL INTEGRITY IN VIVO
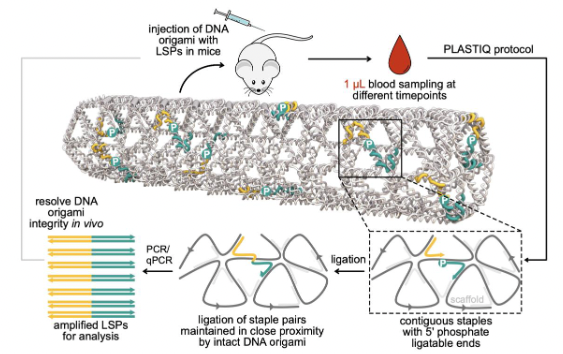
Collaboration with the group of Björn Högberg - a method for tracking the structural integrity of DNA nanostructures in vivo. The principle of the method is to ligate adjacent staple strands, which only works when the structure is intact. Those ligated strands can then by detected with high sensitivity from a drop of blood.
DNA ORIGAMI STRUCTURAL INTEGRITY IN VIVO
Collaboration with the group of Björn Högberg - a method for tracking the structural integrity of DNA nanostructures in vivo. The principle of the method is to ligate adjacent staple strands, which only works when the structure is intact. Those ligated strands can then by detected with high sensitivity from a drop of blood.
Y. Wang, I. Rocamonde Lago, J. Waldvogel, S. Zang, I. Baars, A. Kloosterman, B. Shen, I. T. Hoffecker, Q. He, B. Högberg. DNA Origami Structural Integrity Tracked In VivoUsing Proximity Ligation. Nature Nanotechnology, 2026
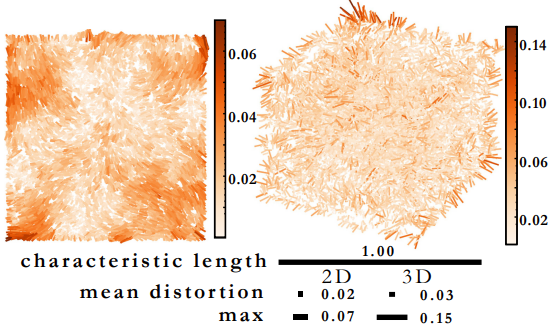 IMAGE RECOVERY FROM UNKNOWN NETWORK MECHANISMS FOR DNA SEQUENCING-BASED MICROSCOPY
In this work, we tackled the problem of how to reconstruct an image from a molecular network formed via unknown mechanisms, a contribution to the emerging field of DNA sequencing-based microscopy. The structure and structure-generating rules of molecular networks need not be known beforehand thanks to efficient use of random walks that we used to perform structural discovery. It turns out this method also saves computing costs, making it both fast and robust.
IMAGE RECOVERY FROM UNKNOWN NETWORK MECHANISMS FOR DNA SEQUENCING-BASED MICROSCOPY
In this work, we tackled the problem of how to reconstruct an image from a molecular network formed via unknown mechanisms, a contribution to the emerging field of DNA sequencing-based microscopy. The structure and structure-generating rules of molecular networks need not be known beforehand thanks to efficient use of random walks that we used to perform structural discovery. It turns out this method also saves computing costs, making it both fast and robust.
D. Fernandez Bonet, I. T. Hoffecker*; Image recovery from unknown network mechanisms for DNA sequencing-based microscopy Nanoscale, 2023

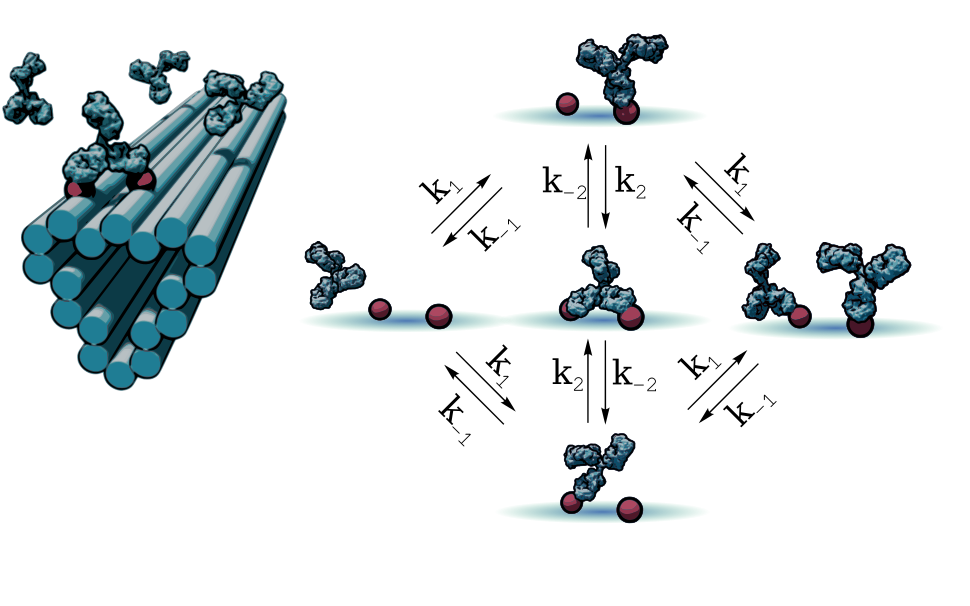 PROGRAMMING ANTIBODY MIGRATION USING PATTERNED ANTIGENS
We took the patterned surface plasmon resonance method further in this work by developing a robust model and parameterization pipeline for antibodies binding to clusters of closely patterned antigens. The most surprising part of this study was the discovery that, due to the spring-like preference of antibodies to bind more stabley to antigen dimers of a particular separation distance, our model actually predicts directed migration of antibodies on antigen patterns with gradients of spacing distance. The model, pipeline, and code can be found here: GitHub
PROGRAMMING ANTIBODY MIGRATION USING PATTERNED ANTIGENS
We took the patterned surface plasmon resonance method further in this work by developing a robust model and parameterization pipeline for antibodies binding to clusters of closely patterned antigens. The most surprising part of this study was the discovery that, due to the spring-like preference of antibodies to bind more stabley to antigen dimers of a particular separation distance, our model actually predicts directed migration of antibodies on antigen patterns with gradients of spacing distance. The model, pipeline, and code can be found here: GitHub
I. T. Hoffecker*, A. Shaw, V. Sorokina, I. Smyrlaki, B. Högberg* ; Stochastic modeling of antibody binding predicts programmable migration on antigen patterns Nature Computational Science, 2022
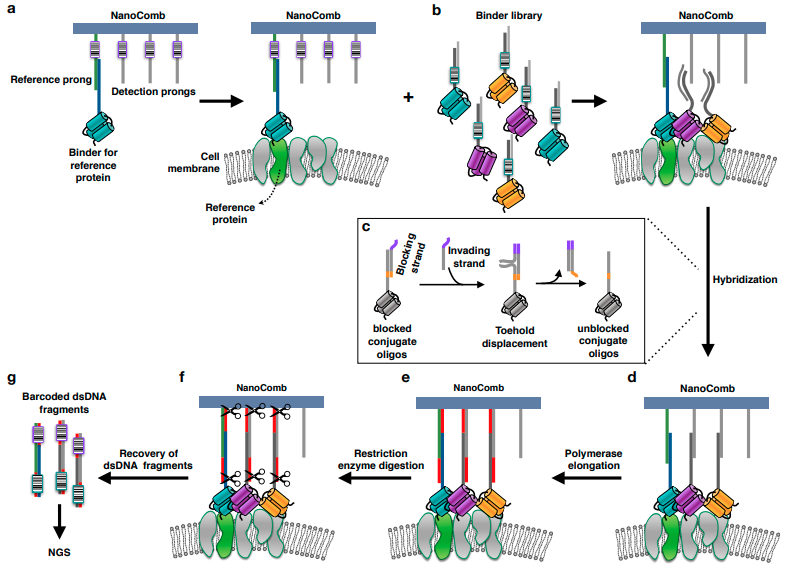 OPTICS-FREE NANOENVIRONMENT MEASUREMENT USING DNA SEQUENCING
This entirely optics-free spatial analysis technique called "NANODEEP" uses DNA probes as molecular recording devices to sample the spatial frequency of nanoscale objects like membrane proteins. Information about object species and relative distance are stored in DNA sequences and recovered using next gen sequencing, revealing the difficult-to-obtain information about the nanoscale spatial distribution of heterogeneous protein clusters. The NANODEEP processing code can be accessed here: GitHub
OPTICS-FREE NANOENVIRONMENT MEASUREMENT USING DNA SEQUENCING
This entirely optics-free spatial analysis technique called "NANODEEP" uses DNA probes as molecular recording devices to sample the spatial frequency of nanoscale objects like membrane proteins. Information about object species and relative distance are stored in DNA sequences and recovered using next gen sequencing, revealing the difficult-to-obtain information about the nanoscale spatial distribution of heterogeneous protein clusters. The NANODEEP processing code can be accessed here: GitHub
E. Ambrosetti, G. Bernardinelli, I. T. Hoffecker, L. Hartmanis, R. Sandberg, B. Högberg, A. I. Teixeira* ; A DNA nanoassembly-based approach to map membrane protein nanoenvironments Nature Nanotechnology, 2021
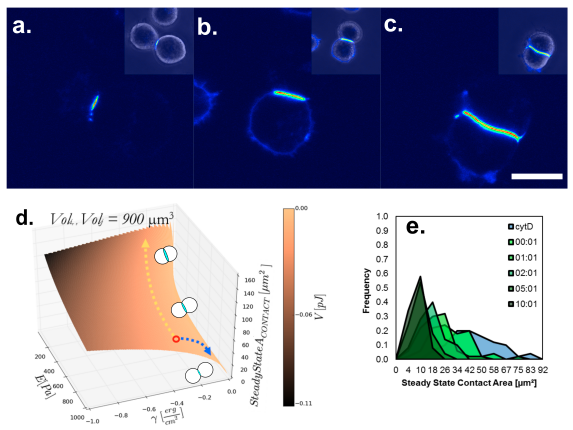 PROGRAMMING THE STRENGTH OF CELL-CELL ADHESION USING DNA
Here, we use DNA-conjugated phospholipid molecules to program heterotypic attachment of cell membranes. Controlling the density of binding molecules enables us to tune the strength of the adhesive interaction of cell-cell doublets. We use this tool to study cell-cell adhesion mechanics as a model that is decoupled from the complex native adhesion machinery in cohering cell types.
PROGRAMMING THE STRENGTH OF CELL-CELL ADHESION USING DNA
Here, we use DNA-conjugated phospholipid molecules to program heterotypic attachment of cell membranes. Controlling the density of binding molecules enables us to tune the strength of the adhesive interaction of cell-cell doublets. We use this tool to study cell-cell adhesion mechanics as a model that is decoupled from the complex native adhesion machinery in cohering cell types.
I. T. Hoffecker*, Y. Arima, H Iwata; Tuning intercellular adhesion with membrane-anchored oligonucleotides J. Royal Soc. Interface, 2019

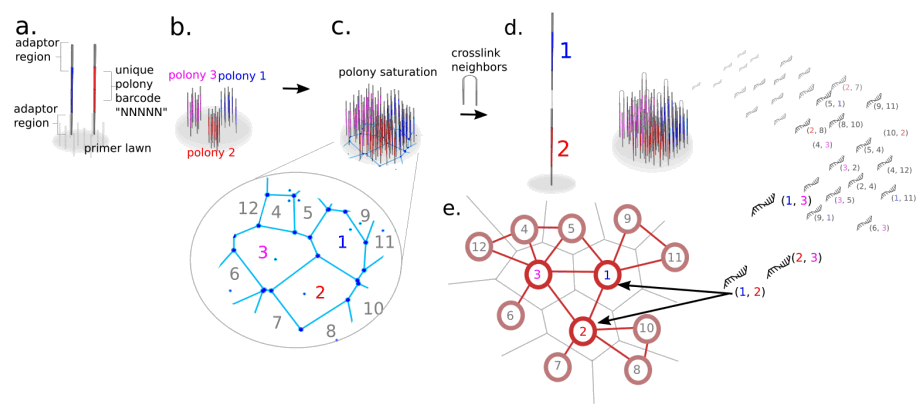 OPTICS-FREE MICROSCOPY USING DNA SEQUENCING
The idea behind this project is to invent a new kind of microscopy that uses DNA sequences instead of light as the primary medium of information transmission. The paper is a computational proof of concept that a dense saturated surface of polonies could be used to store spatial information on a surface that could then be reconstructed into whole images after sequencing and computational processing.
OPTICS-FREE MICROSCOPY USING DNA SEQUENCING
The idea behind this project is to invent a new kind of microscopy that uses DNA sequences instead of light as the primary medium of information transmission. The paper is a computational proof of concept that a dense saturated surface of polonies could be used to store spatial information on a surface that could then be reconstructed into whole images after sequencing and computational processing.
I.T. Hoffecker, Y. Yang, G. Bernardinelli, P. Orponen, B. Högberg* ; A Computational Framework for DNA Sequencing-Based Microscopy PNAS, 2019
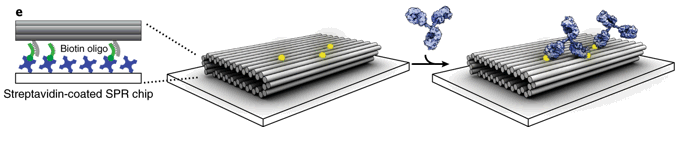
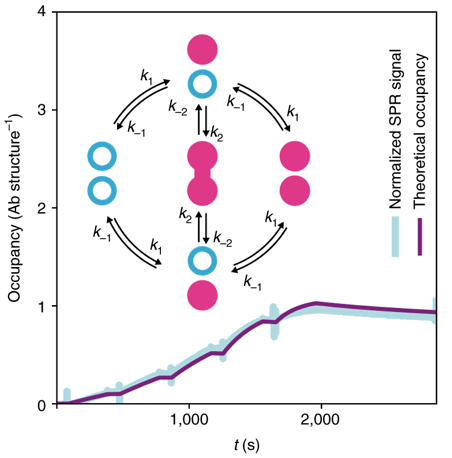 MULTIVALENT ANTIBODY/ANTIGEN BINDING KINETICS WITH DNA ORIGAMI
Using DNA origami, it was possible to pattern and modulate spacing of antigens in a controlled reproducible manner. We could then examine the multivalent binding kinetics of antibodies with surface plasmon resonance like never before. My role in the paper was to model the kinetics, calibrate it with real data (left), and generate estimates of the underlying distribution of microstates comprising each point in time.
MULTIVALENT ANTIBODY/ANTIGEN BINDING KINETICS WITH DNA ORIGAMI
Using DNA origami, it was possible to pattern and modulate spacing of antigens in a controlled reproducible manner. We could then examine the multivalent binding kinetics of antibodies with surface plasmon resonance like never before. My role in the paper was to model the kinetics, calibrate it with real data (left), and generate estimates of the underlying distribution of microstates comprising each point in time.
A. Shaw, I.T. Hoffecker, I. Smyrlaki, J. Rosa, A. Grevys, D. Bratlie, I. Sandlie, T. E. Michaelsen, J. T. Andersen, B. Högberg*; Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies Nature Nanotechnology, 2019
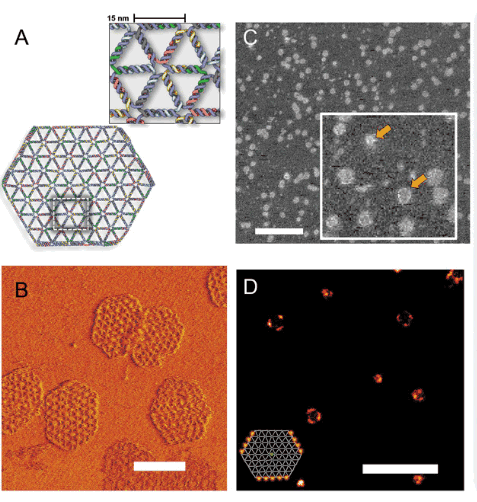 USING SOLUTIONS TO TOGGLE OPENING AND CLOSING OF A DNA NANOSTRUCTURE
By manipulating solution conditions, we could open and close a tethered DNA nanostructure. We used DNA PAINT and image classification to characterize the transitions.
USING SOLUTIONS TO TOGGLE OPENING AND CLOSING OF A DNA NANOSTRUCTURE
By manipulating solution conditions, we could open and close a tethered DNA nanostructure. We used DNA PAINT and image classification to characterize the transitions.
I.T. Hoffecker, S. Chen, A. Gådin, A. Bosco, A. I. Teixeira*, B. Högberg* ; Solution‐Controlled Conformational Switching of an Anchored Wireframe DNA Nanostructure Small, 2018
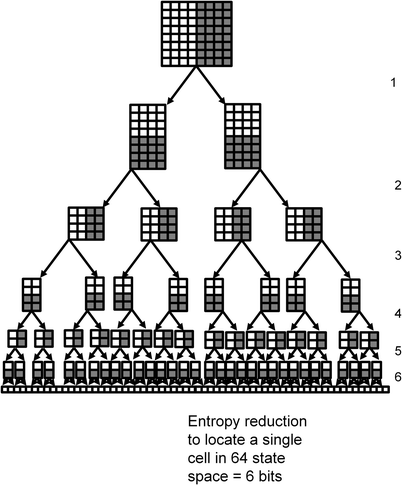 ENTROPY, COMPLEXITY, AND FUNCTION IN EARLY HUMAN TECHNOLOGY
Structural organization has always been used to extract inferences about the function of biological phenomena. We take a formal approach to this idea with different metrics of complexity to characterize early the technology of early humans and discuss how this sheds light on the problems they faced.
ENTROPY, COMPLEXITY, AND FUNCTION IN EARLY HUMAN TECHNOLOGY
Structural organization has always been used to extract inferences about the function of biological phenomena. We take a formal approach to this idea with different metrics of complexity to characterize early the technology of early humans and discuss how this sheds light on the problems they faced.
J. F. Hoffecker, I.T. Hoffecker; The Structural and Functional Complexity of Hunter-Gatherer Technology Journal of Archaeological Method and Theory, 2018
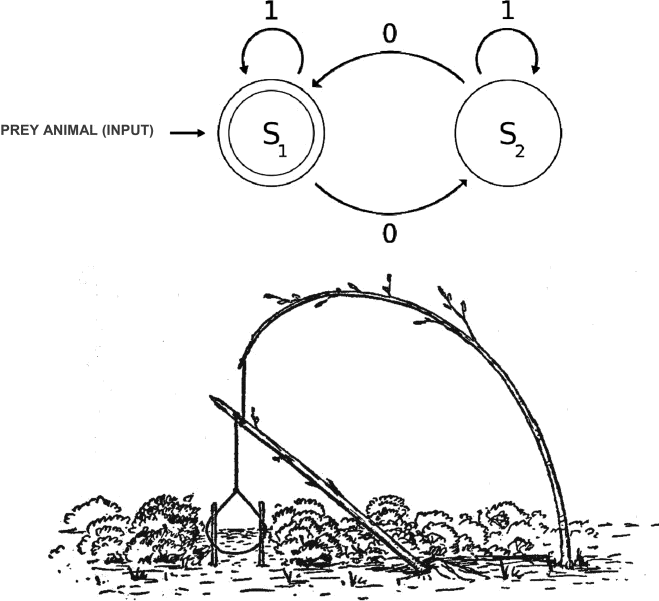 APPLYING INFORMATION THEORY TO HUMAN EVOLUTION
Information, automata, and computation theory have shed light on nearly every corner of biological science...except archaeology. That is, until now! In this paper we examine the global dispersal of modern humans through the lens of computational complexity as it applies to early human technologies.
APPLYING INFORMATION THEORY TO HUMAN EVOLUTION
Information, automata, and computation theory have shed light on nearly every corner of biological science...except archaeology. That is, until now! In this paper we examine the global dispersal of modern humans through the lens of computational complexity as it applies to early human technologies.
J. F. Hoffecker, I.T. Hoffecker; Technological complexity and the global dispersal of modern humans Evolutionary Anthropology: Issues, News, and Reviews, 2017
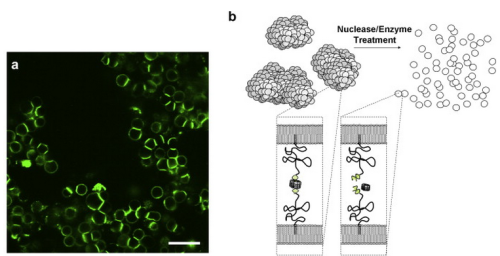
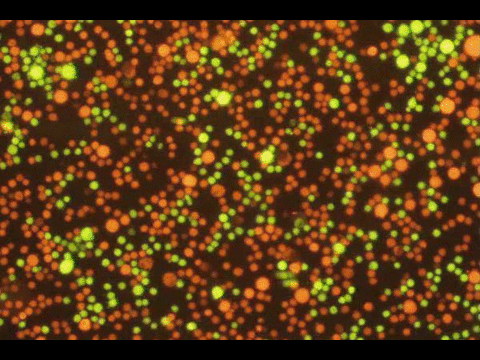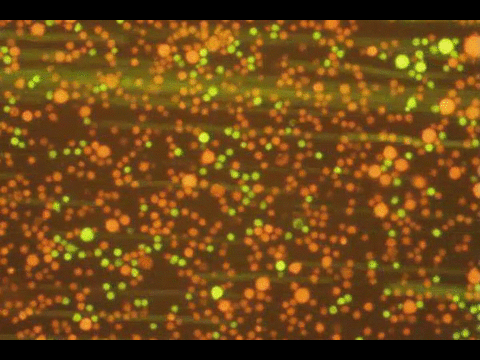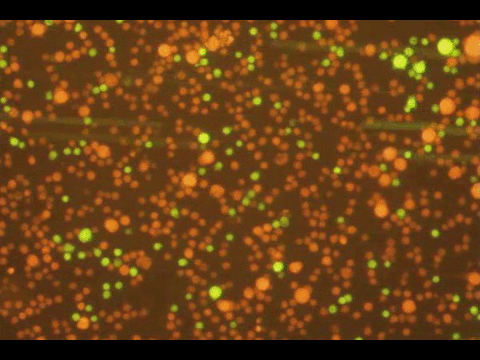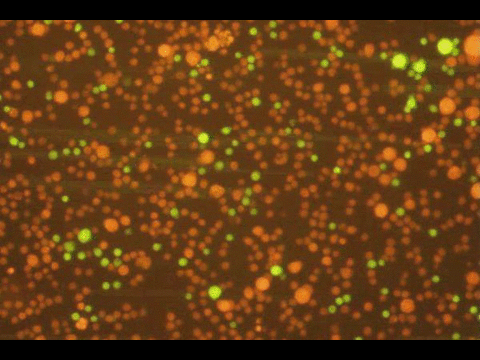 PROGRAMMABLE CELL ATTACHMENT AND SEVERING
In this paper, we explore methods to cleave DNA mediating artificial attachment of DNA-lipid-modified cells to flat substrates or other cells. We used BamHI to sever the DNA tethers of cells bearing the correct BamHI recognition site while leaving other tethered cells alone. We also used Benzonase, a powerful nonspecific nuclease, to digest all the tethers and detach cells irrespective of their anchor sequences. Now we have a way to programmably attach and detach cells!
PROGRAMMABLE CELL ATTACHMENT AND SEVERING
In this paper, we explore methods to cleave DNA mediating artificial attachment of DNA-lipid-modified cells to flat substrates or other cells. We used BamHI to sever the DNA tethers of cells bearing the correct BamHI recognition site while leaving other tethered cells alone. We also used Benzonase, a powerful nonspecific nuclease, to digest all the tethers and detach cells irrespective of their anchor sequences. Now we have a way to programmably attach and detach cells!
I.T. Hoffecker, N. Takemoto, Y. Arima, H. Iwata*; Sequence-specific nuclease-mediated release of cells tethered by oligonucleotide phospholipids Biomaterials, 2015
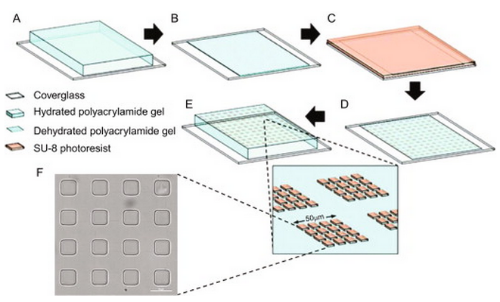 PATTERNING SOFT GELS WITH STIFF ISLANDS
To examine the rigidity sensing mechanisms of cells, I previously designed a method for producing micropatterned arrays of rigid photoresist islands grafted to an underlying hydrogel of tunable stiffness. The islands can be made adhesive to cells while the exposed hydrogel is left inert. This is the detailed protocol for producing such substrates.
PATTERNING SOFT GELS WITH STIFF ISLANDS
To examine the rigidity sensing mechanisms of cells, I previously designed a method for producing micropatterned arrays of rigid photoresist islands grafted to an underlying hydrogel of tunable stiffness. The islands can be made adhesive to cells while the exposed hydrogel is left inert. This is the detailed protocol for producing such substrates.
S. Wong, W.H. Guo, I.T. Hoffecker, Y.L. Wang*; Preparation of a micropatterned rigid-soft composite substrate for probing cellular rigidity sensing Methods Cell Biol, 2014
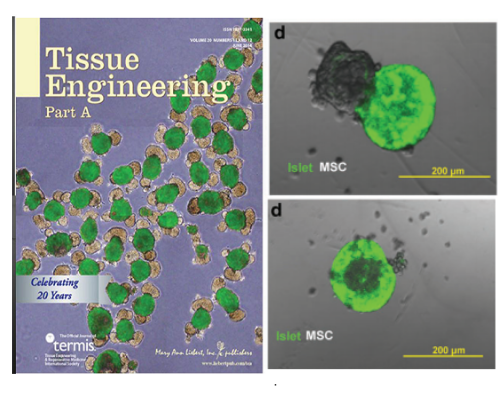
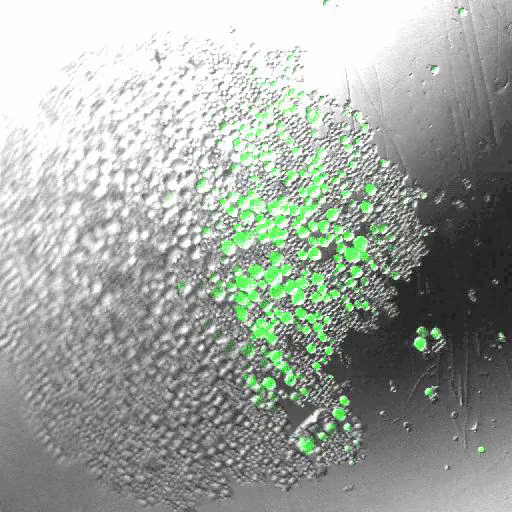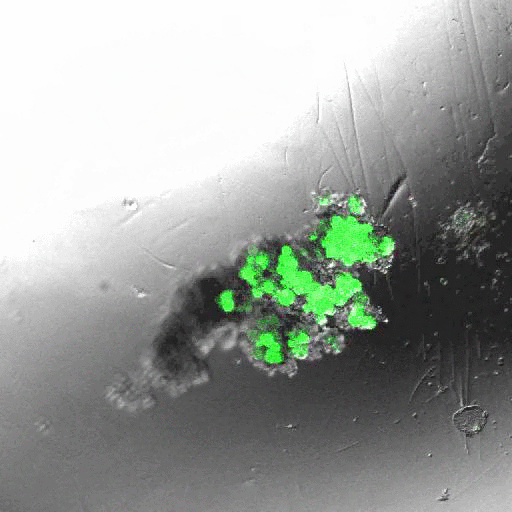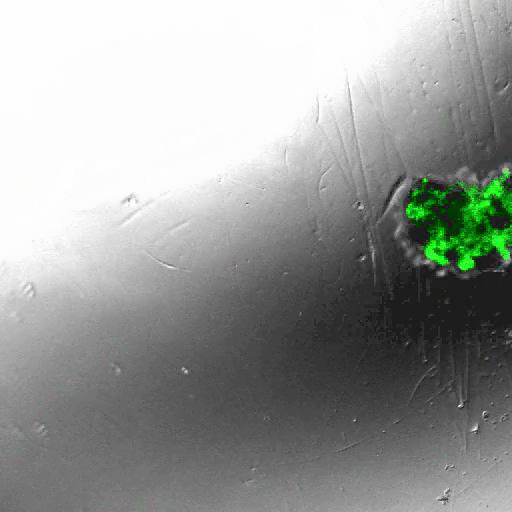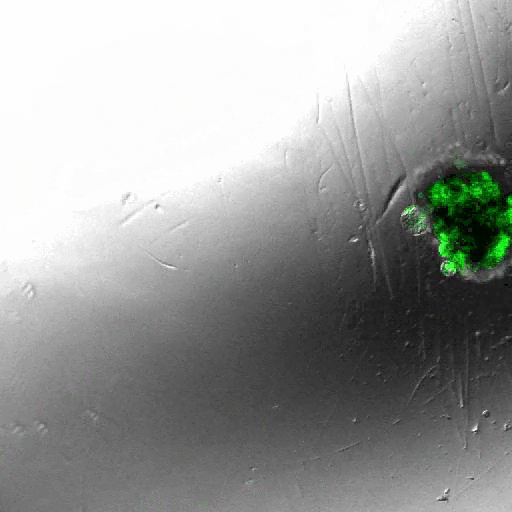 INDUCING MAJOR CHANGES IN CELL SORTING PATTERNS
Cell sorting is a process where cells of different types spontaneously undergo relative migration, leading to distinct multicellular domains of differing cell type composition. The range of cell-cell communication is limited, and global patterns emerge due to the iteration of interactions between neighbors. We observed major changes in global sorting patterns after pharmacologically inducing changes in the individual cell-cell cohesive interactions.
INDUCING MAJOR CHANGES IN CELL SORTING PATTERNS
Cell sorting is a process where cells of different types spontaneously undergo relative migration, leading to distinct multicellular domains of differing cell type composition. The range of cell-cell communication is limited, and global patterns emerge due to the iteration of interactions between neighbors. We observed major changes in global sorting patterns after pharmacologically inducing changes in the individual cell-cell cohesive interactions.
I.T. Hoffecker, H. Iwata*; Manipulation of cell sorting in mesenchymal stromal cell-islet cell co-aggregate spheroids Tissue Engineering Part A, 2014
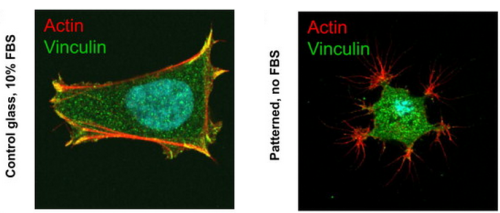 DNA-LIPID-MEDIATED ADHESION vs INTEGRIN-MEDIATED ADHESION
We induced an artificial form of adhesion between cells modified with ssDNA-lipids and substrates inkjet-printed with complementary ssDNA. When incubated without serum or adhesive proteins in the media, cells took on a bizarre dendrite-like morphology. When in the presence of serum proteins, cells slowly pulled away from their DNA anchors leaving trails of lipid matter behind. This study demonstrates the clear and interesting distinction between natural and artificially induced forms of adhesion.
DNA-LIPID-MEDIATED ADHESION vs INTEGRIN-MEDIATED ADHESION
We induced an artificial form of adhesion between cells modified with ssDNA-lipids and substrates inkjet-printed with complementary ssDNA. When incubated without serum or adhesive proteins in the media, cells took on a bizarre dendrite-like morphology. When in the presence of serum proteins, cells slowly pulled away from their DNA anchors leaving trails of lipid matter behind. This study demonstrates the clear and interesting distinction between natural and artificially induced forms of adhesion.
K. Sakurai, I.T. Hoffecker, H. Iwata*; Long term culture of cells patterned on glass via membrane-tethered oligonucleotides Biomaterials, 2013
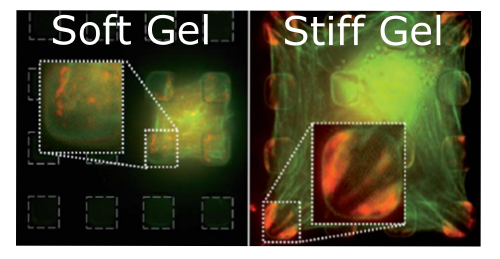 CELL RIGIDITY SENSING IS BLIND TO FINE DETAILS
Do cell focal adhesion contain molecular mechanical sensors to measure the intrinsic stiffness of their underlying substrates? Perhaps not. We built micropatterned arrays of rigid islands of sub-cellular size grafted to a soft underlying hydrogel. Though their adhesive contact was restricted to the islands, the cells on these patterns behaved as they would on uniformly soft substrates, apparently ignoring the intrinsic stiffness of the islands. You could say that cells 'pull' their substrates rather than 'pinching' them.
CELL RIGIDITY SENSING IS BLIND TO FINE DETAILS
Do cell focal adhesion contain molecular mechanical sensors to measure the intrinsic stiffness of their underlying substrates? Perhaps not. We built micropatterned arrays of rigid islands of sub-cellular size grafted to a soft underlying hydrogel. Though their adhesive contact was restricted to the islands, the cells on these patterns behaved as they would on uniformly soft substrates, apparently ignoring the intrinsic stiffness of the islands. You could say that cells 'pull' their substrates rather than 'pinching' them.
I.T. Hoffecker, W.H. Guo, Y.L. Wang*; Assessing the spatial resolution of cellular rigidity sensing using a micropatterned hydrogel-photoresist composite Lab on a Chip, 2011
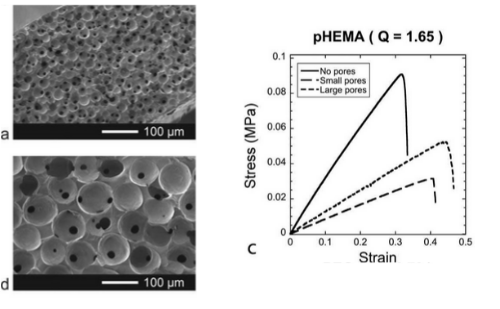 MECHANICS OF POROUS HYDROGEL SCAFFOLDS
Preparing hydrogel scaffolds with pores via sphere-templating reduced the elastic modulus and ultimate tensile stress but interestingly increased ultimate tensile strain. We looked at several parameters such as polymer type, pore diameter, and gel crosslinking density and examined their effect on scaffold mechanical properties. Our results will help guide better hydrogel scaffold designs for tissue engineering.
MECHANICS OF POROUS HYDROGEL SCAFFOLDS
Preparing hydrogel scaffolds with pores via sphere-templating reduced the elastic modulus and ultimate tensile stress but interestingly increased ultimate tensile strain. We looked at several parameters such as polymer type, pore diameter, and gel crosslinking density and examined their effect on scaffold mechanical properties. Our results will help guide better hydrogel scaffold designs for tissue engineering.
S.M. LaNasa, I.T. Hoffecker, S.J. Bryant*; The role of pore size on the mechanical properties of porous poly(ethylene glycol) and poly(2-hydroxyethyl methacrylate) hydrogels Journal of Biomedical Materials Research - Part B: Applied Biomaterials, 2011