Understanding Spatial Networks
Spatial networks are a simple but powerful way to describe how things are arranged in space when we do not have a map or coordinates to begin with.
Instead of recording where objects are, we only record which ones are connected or close to each other. These connections might represent wireless communication between sensors,
train routes between cities, or physical interactions between molecules.
In such a network, the objects are called nodes and the connections between them are called edges.
When edges tend to link nearby objects rather than distant ones, the network quietly contains information about space.
A central question we study is how much of that hidden spatial information can be recovered from the network alone,
and what properties make a network truly informative about geometry rather than just connectivity.
← Click!
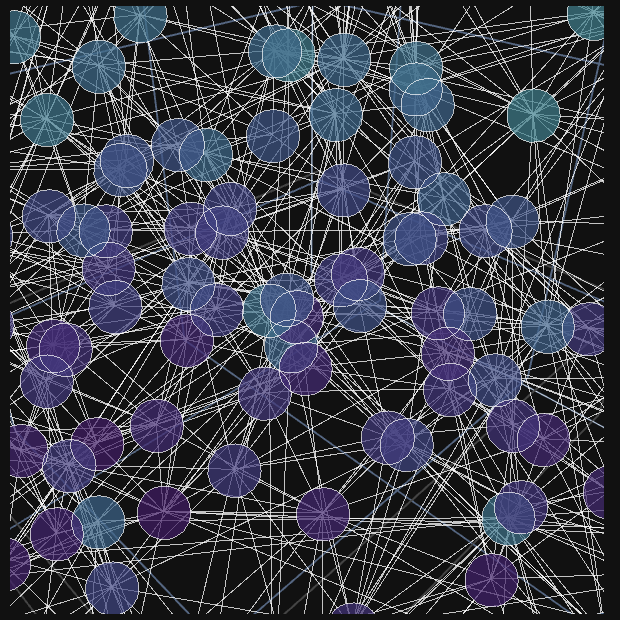
In the interactive panel here, each point (node) represents a molecule, and each line (edge) represents a recorded interaction between nearby molecules.
If you zoom into the cloud of nodes and edges, you can see that edges tend to connect spatial neighbors.
This means that the network structure is correlated with physical proximity. Even when this correlation is imperfect,
it can still be used to infer spatial organization directly from the network.
Click! →
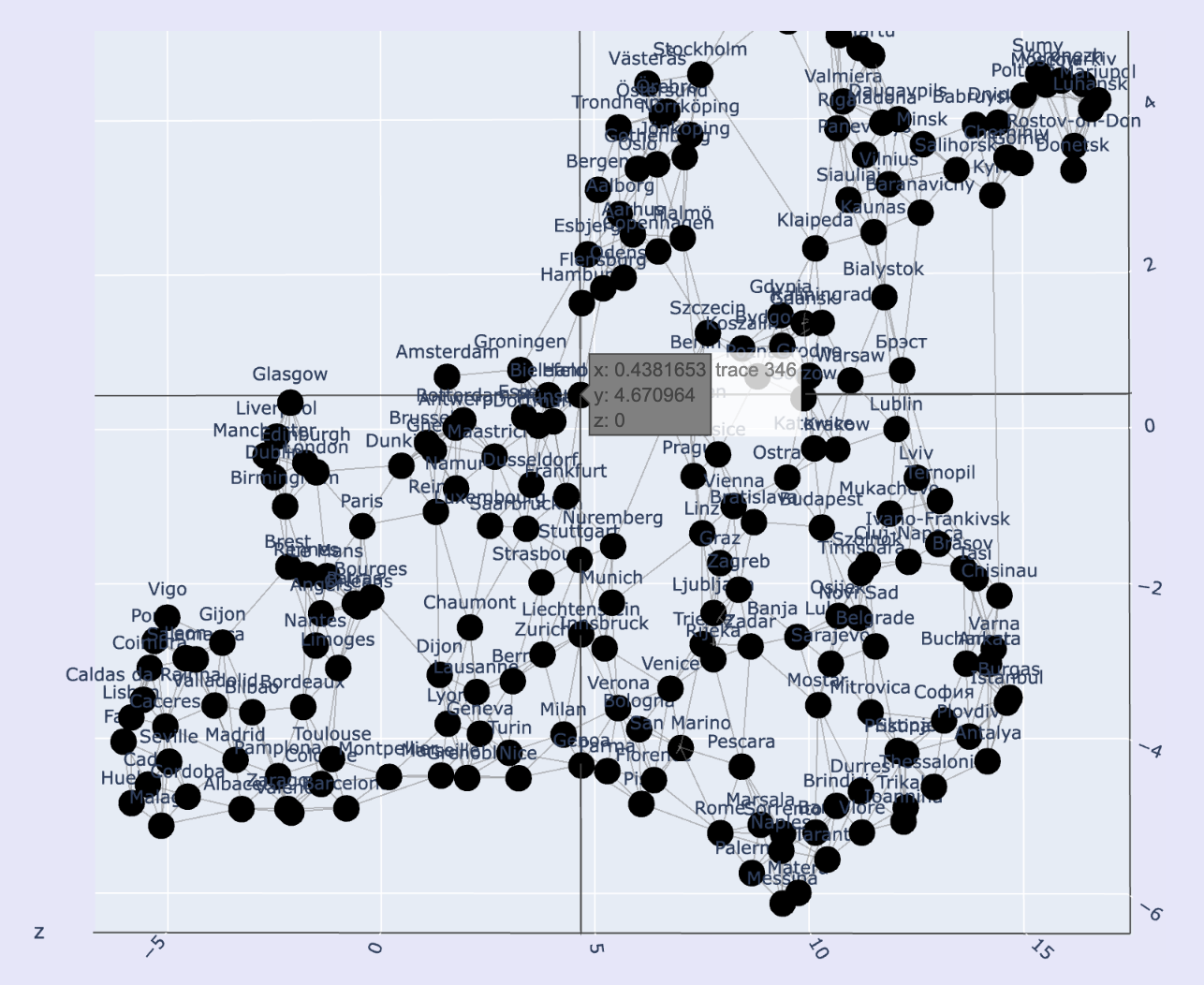
A big list of regional transit connections in Europe, e.g. Stockholm to Copenhagen or Paris to Lyon, constitutes a spatial network. A network is "spatial" if connections are related to the distance between objects.
So a list of regional transit connections in fact gives us information about where, in space, the cities are located relative to one another.
By optimizing for satisfaction of constraints, i.e. maximizing the closeness of connected nodes, we can recover that spatial information and generate an approximation -
have a look at the interactive plot here. -> We can see a few things - first of all the global positions have not be preserved.
However try reflecting and rotate this image, and you should be able to arrive at a more familiar map of Europe.
Remember that no information about coordinates was passed through to this reconstruction.
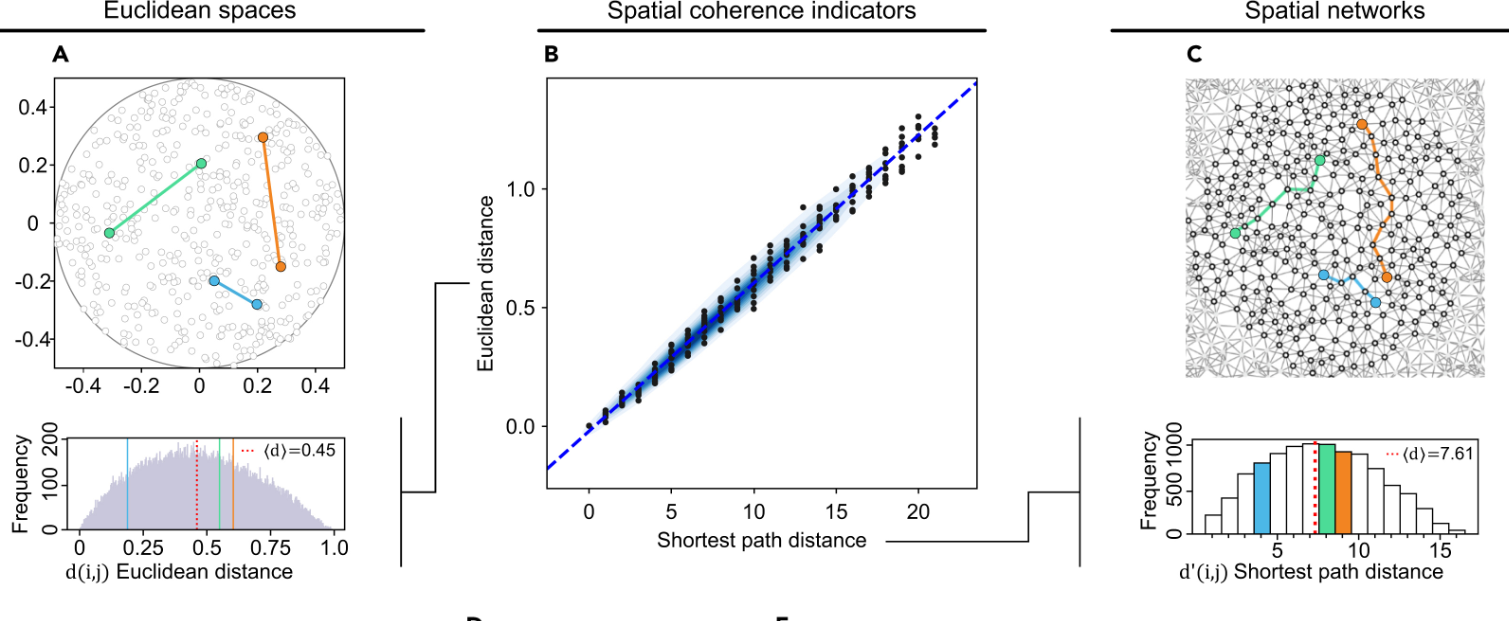
Understanding spatial networks addresses a core problem shared across biology, physics, and data science: how reliable geometric information can emerge from local interactions alone.
Many natural and engineered systems do not provide direct access to coordinates, but instead encode space implicitly through patterns of connectivity, proximity, or interaction.
Our work focuses on identifying the principles that make such networks reconstructable, asking when and why a network genuinely reflects an underlying physical or
geometric space rather than an abstract graph. Central to this is the concept of spatial coherence, which captures whether network distances behave consistently with
geometric constraints such as dimensionality, scaling, and geodesic structure.
By studying how shortest path distances, spectral properties, and scaling laws relate to physical space, we seek to distinguish networks that preserve spatial meaning
from those distorted by noise, shortcuts, or missing information.
Beyond reconstruction itself, spatial networks raise broader questions about robustness, noise tolerance, and design. Real networks often contain false or long range connections, uneven sampling, or heterogeneous density, all of which can blur spatial structure. Rather than treating these effects as purely detrimental, we study how network properties such as connectivity and redundancy influence the balance between signal and distortion. This perspective allows us to develop ground truth free quality control measures and principled ways to assess whether a dataset is likely to support reliable spatial inference. More generally, our research aims to establish a theory of spatial networks that applies across modalities, from molecular and cellular systems to sensor networks and abstract embeddings, providing tools to reason about space when space itself is not directly observed.
Related Publications & Links:
- Bonet DF, Blumenthal JI, Lang S, Dahlberg SK, Hoffecker IT. Spatial coherence in DNA barcode networks. Patterns. 2025 Dec 12;6(12).
cell.com
- Bonet DF, Hoffecker IT. Image recovery from unknown network mechanisms for DNA sequencing-based microscopy. Nanoscale. 2023;15(18):8153-7.
pubs.rsc.org
- Hoffecker IT, Yang Y, Bernardinelli G, Orponen P, Högberg B. A computational framework for DNA sequencing microscopy. Proceedings of the National Academy of Sciences. 2019 Sep 24;116(39):19282-7.
pnas.org
Network-based 2D Spatial Transcriptomics
Spatial transcriptomics is a set of technologies that makes it possible to measure which genes are active in a
tissue while preserving their physical locations. This matters because biological function is not determined by gene
activity alone, but by how different cell types are arranged, interact, and form structures such as layers, gradients,
and microenvironments. By adding spatial context to gene expression measurements, spatial transcriptomics allows researchers
to study tissue organization in development, cancer, and disease, and to connect molecular programs to anatomy in a direct
and quantitative way.
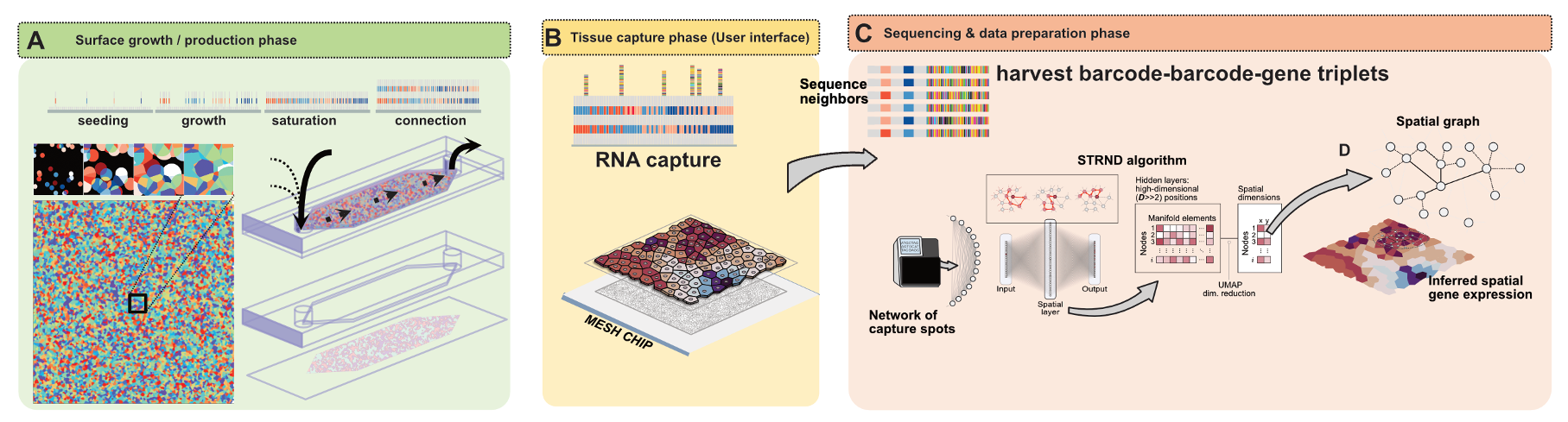
Network based 2D spatial transcriptomics is an emerging class of approaches that encodes spatial information implicitly through molecular
proximity rather than explicitly through pre defined coordinates.
Instead of relying on printed grids, lithography, or optical decoding to assign positions,
these methods generate dense collections of molecular capture sites whose local neighborhood relationships are recorded in sequencing data. By reading out which molecular barcodes tend to appear together due to physical proximity on a surface, one obtains a graph whose structure reflects the underlying tissue geometry.
Spatial positions are then inferred computationally by reconstructing a two dimensional layout that is most consistent with the observed network connectivity,
in close analogy to recovering a map from a list of local connections.
This network based perspective shifts spatial transcriptomics from a manufacturing problem to an inference problem.
Spatial resolution and mapped area are no longer fixed by the precision of fabrication, but instead emerge from the density and quality of local connections
in the network and from the reconstruction algorithms applied downstream. As a result, these approaches offer a path toward large area, high throughput spatial
profiling using simple, self assembled capture surfaces combined with standard sequencing. More broadly, network based spatial transcriptomics connects molecular
biology with graph theory and statistical inference, opening new opportunities to study tissue organization, gradients, and multicellular structure at scale while
reducing cost and technical complexity.
Related Publications & Links:
- Benson E, Hoffecker IT. Capture surface of metapolonies. Swedish Patent SE547614C2. Application filed January 15, 2024. Published October 28, 2025.
patents.google.com
- Hoffecker IT, Yang Y, Bernardinelli G, Orponen P, Högberg B. A computational framework for DNA sequencing microscopy. Proceedings of the National Academy of Sciences. 2019 Sep 24;116(39):19282-7.
pnas.org
- Two researchers in Sweden to receive an ERC Proof of Concept Grant
www.vr.se
- Spatial transcriptomics chips with sequencing-based microscopy
cordis.europa.eu
- Dahlberg SK, Bonet DF, Franzén L, Ståhl PL, Hoffecker IT. Hidden network preserved in Slide-tags data allows reference-free spatial reconstruction. Nature Communications. 2025 Oct 31;16(1):9652.
nature.com
3D Molecular Reconstruction by Sequencing-based Microscopy

Three dimensional molecular imaging aims to reveal how cells, genes, and biomolecules are organized and interact inside
intact tissues and organs. This spatial context is essential for understanding development, disease progression, and how
complex biological functions emerge from local interactions. Today, most high resolution 3D imaging relies on optical microscopy,
which faces fundamental challenges as samples become thicker and more complex. Light scattering, absorption, limited penetration depth,
and the need for elaborate clearing or sectioning protocols constrain resolution, throughput, and scalability. As a result, many biological
systems remain difficult or impossible to image comprehensively in three dimensions using optics alone.
Our research explores an alternative route to 3D molecular imaging that does not rely on lenses or light. Instead of measuring spatial
position directly, we encode spatial proximity into networks of interacting molecular barcodes. When molecules are close together in
physical space, they are more likely to interact, co associate, or be captured together. By reading out these interactions using high
throughput DNA sequencing, we obtain large molecular networks whose connectivity reflects the underlying three dimensional structure of the
sample. In this framework, spatial information is stored implicitly in who is connected to whom, rather than explicitly in pixels or voxels.
From these molecular networks, we reconstruct three dimensional spatial organization computationally. Using graph based inference,
we recover layouts that are most consistent with the observed connectivity patterns, effectively rebuilding a 3D map from local neighborhood
relationships alone. This network based, optics free approach shifts 3D microscopy from a hardware limited imaging problem to a data driven
inference problem. It opens a path toward scalable, tissue scale, and potentially whole organ molecular imaging using sequencing as the primary
readout, with resolution and volume determined by network density and molecular design rather than by optical constraints.
Related Publications & Links:
- Bonet DF, Blumenthal JI, Lang S, Dahlberg SK, Hoffecker IT. Spatial coherence in DNA barcode networks. Patterns. 2025 Dec 12;6(12).
cell.com
- Bonet DF, Hoffecker IT. Image recovery from unknown network mechanisms for DNA sequencing-based microscopy. Nanoscale. 2023;15(18):8153-7.
pubs.rsc.org
- Instrument-free 3D molecular imaging with the VOLumetric UMI-Network EXplorer
cordis.europa.eu
- VOLUMINEX Project Webpage
voluminex-project.eu/
- SciLifeLab-led consortium receives Pathfinder grant to enable sequencing-based microscopy in 3D
scilifelab.se
- KTH med och delar på 11,7 miljoner euro till deep tech
kth.se
- The Future of 3-D Molecular Imaging in Life Sciences with Ian Hoffecker
singletechnologies.com
Technology & Evolutionary Computation
Technology can be understood as a form of computation carried out in the physical world. Every technological process encodes rules, constraints, and memory, and unfolds through sequences of conditional actions, much like a program being executed. Over time, technological designs change through processes that closely resemble biological evolution: variants are generated, tested through interaction with their environment, and selectively retained. Innovation in this view is not a single moment of invention, but an evolutionary process shaped by variation, selection, and inheritance. This perspective allows technologies, from molecular systems to large-scale material practices, to be studied as evolving populations governed by general principles of information processing.
Our research explores these principles at a fundamental level by developing formal models of technology as a generative process. We study how structured designs arise from sequences of dependent operations, how techniques and subcomponents are reused across different constructions, and how material outcomes preserve only partial information about the processes that produced them. By framing technological production as a stochastic, hierarchical process with hidden internal organization, we aim to understand both how complex technological systems emerge and what can be inferred about them from incomplete physical evidence.
These ideas also inform our interest in evolutionary computation as a design strategy in molecular programming. Through selected collaborations, we engage with systems where variation and selection are used to explore functional design spaces, such as DNA-based molecular binders shaped through iterative selection. In this context, evolutionary processes serve as both a conceptual model and a practical tool, illustrating how computation, adaptation, and physical implementation can coincide. This research theme situates evolutionary computation as a unifying lens for understanding technology across scales, without treating any single application domain as its primary focus.
Related Publications & Links:
- Benson E, Hoffecker IT. Random DNA structures. International Patent Application WO2024260866. Application filed June 14, 2024. Published December 26, 2024.
patentscope.wipo.int
- Hoffecker JF, Hoffecker IT. Technological complexity and the global dispersal of modern humans. Evolutionary Anthropology: Issues, News, and Reviews. 2017 Nov;26(6):285-99.
onlinelibrary.wiley.com
- Hoffecker JF, Hoffecker IT. Measuring the computational complexity of artifact design in Paleolithic archaeology.
researchgate.net
- Hoffecker JF, Hoffecker IT. The structural and functional complexity of hunter-gatherer technology. Journal of Archaeological Method and Theory. 2018 Mar;25(1):202-25.
link.springer.com
Spatially Programmed Immunochemistry
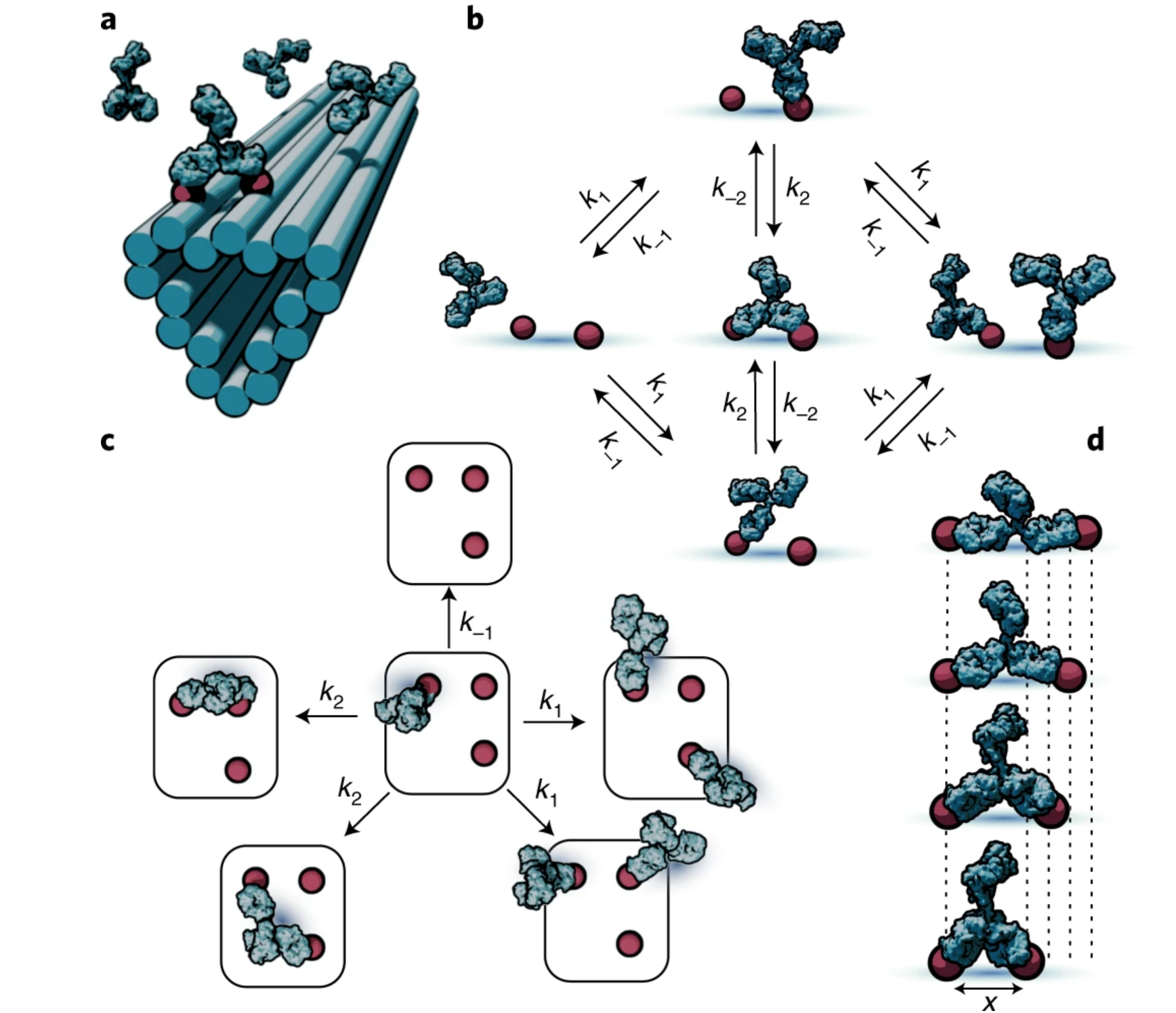
At the most basic level, this research asks a simple question:
how does the immune system sense and respond to the spatial
arrangement of molecules, not just their chemical identity?
In living systems, many biological signals are encoded in patterns.
Viruses, bacteria, and synthetic vaccines often present antigens
in highly organized nanoscale arrays, and immune receptors do not
bind these patterns passively. Instead, they physically explore them.
By creating precisely controlled molecular landscapes,
we can study how antibodies move, bind, and cooperate across space,
revealing rules that are otherwise hidden in complex biological environments.
To access these rules, we use DNA nanotechnology as a molecular
construction toolkit. DNA origami allows us to position antigens
with nanometer precision and to systematically vary their spacing
and geometry. By combining these engineered patterns with
quantitative biophysical measurements, we can directly measure
how antibody binding strength, flexibility, and multivalent
interactions depend on spatial organization. This approach has
shown that antibody binding is not governed by a single optimal
distance, but instead reflects a range of spatial tolerances that
depend on antibody class, affinity, and geometry. In this way,
spatial patterning becomes a controllable variable rather than an
uncontrolled feature of biology .
More broadly, this work connects to our group’s central theme
of molecular programming. Rather than viewing molecules as
isolated actors, we treat them as components in a programmable
system, where function emerges from arrangement, connectivity,
and interaction rules. By designing molecular patterns that
encode specific spatial and kinetic constraints, we effectively
write programs that antibodies and other biomolecules execute
through physical interactions. This perspective links immune
recognition to a wider framework of molecular computation,
where nanoscale structure serves as both information storage
and instruction set, guiding biological behavior through
designed molecular geometry.
Related Publications & Links:
- Shaw A, Hoffecker IT, Smyrlaki I, Rosa J, Grevys A, Bratlie D, Sandlie I, Michaelsen TE, Andersen JT, Högberg B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nature nanotechnology. 2019 Feb;14(2):184-90.
nature.com
- Hoffecker IT, Shaw A, Sorokina V, Smyrlaki I, Högberg B. Stochastic modeling of antibody binding predicts programmable migration on antigen patterns. Nature computational science. 2022 Mar;2(3):179-92.
nature.com
- Model shows how antibodies navigate pathogen surfaces like a child at play
eurekalert.org
- DNA origami: A precise measuring tool for optimal antibody effectiveness
sciencedaily.com
DNA-based Chemical Neural Networks
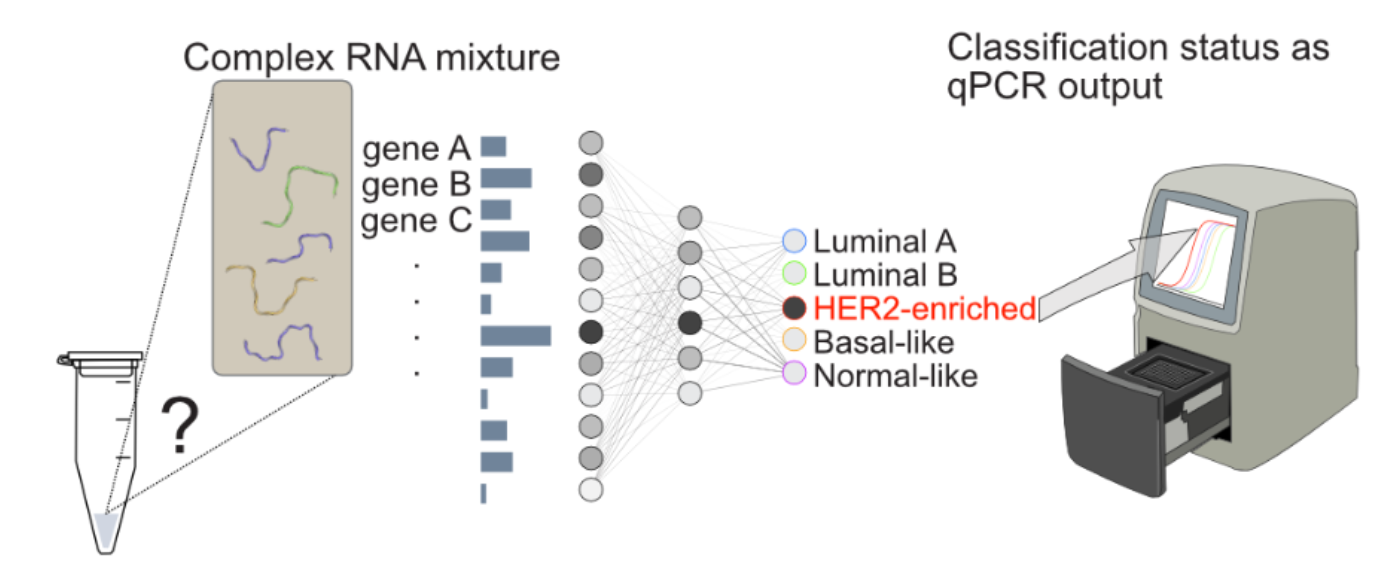
Living cells already process information using chemistry: molecules bind,
react, and amplify signals to make decisions such as when to divide,
differentiate, or respond to stress. DNA-based computing takes inspiration
from this natural logic and asks whether we can deliberately program
molecules to carry out information processing tasks. Instead of electrons
flowing through silicon circuits, information is represented by the
presence and relative amounts of DNA strands, and computation happens
through predictable chemical interactions between them. This opens the
possibility of performing complex decision making directly inside a test
tube, using the same molecular language as biology itself.
In our work, we explore how concepts from artificial neural networks
can be translated into DNA chemistry. In a neural network, information
flows through weighted connections and nonlinear activation steps to
recognize patterns rather than single signals. We recreate these ideas
using DNA sequence design and enzymatic reactions. The strength of
interaction between different DNA strands plays a role similar to
connection weights, while polymerase-driven amplification introduces
nonlinear responses analogous to neural activation. By carefully
designing these molecular interactions, we can build chemical systems
that respond to combinations of inputs, not just the presence or absence
of one molecule.
This approach is especially powerful for diagnosing complex diseases
such as breast cancer, where clinically relevant information is often
encoded in patterns of gene expression rather than single biomarkers.
Today, recognizing these patterns typically requires sequencing and
heavy computational analysis. Our goal is to shift part of that
intelligence into chemistry itself. By constructing DNA-based neural
networks that directly process mixtures of nucleic acids associated
with disease states, we aim to create fast, low-cost diagnostic
reactions that output an interpretable molecular signal. In the
long term, this could help bring sophisticated pattern-based diagnostics
closer to the patient, without the need for centralized sequencing
infrastructure.
Digital Data Storage in DNA
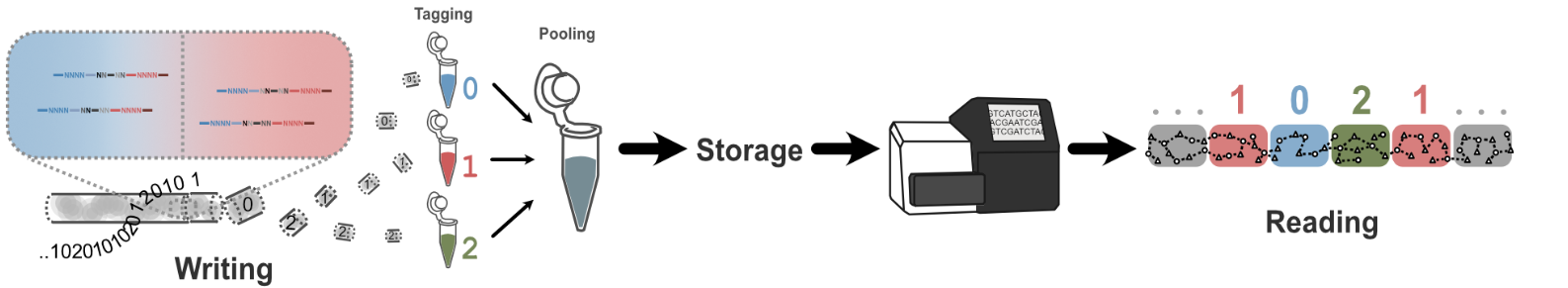
Dna has emerged as an intriguing medium for long-term information storage.
It is extraordinarily dense, stable over geological timescales, and already
manufactured and copied at massive scale by biology.
However, translating these advantages into a practical data storage technology
is still challenging.
Current approaches face bottlenecks related to writing cost and speed
and the difficulty of scaling.
Many of these limitations arise because dna data storage has often been treated as
a linear pipeline. Information is encoded into sequences, synthesized, stored,
and later sequenced and decoded, with each step optimized largely in isolation.
This view struggles with issues such as uneven sequence usage, loss of molecules,
amplification bias, and the growing overhead required for error correction and
indexing as datasets become larger. As a result, there is a widening gap between
elegant proof of concept studies and storage architectures that could operate
robustly in realistic settings.
Our research explores an alternative perspective that treats DNA data
storage as a distributed and structured system rather than a
collection of independent strands. Instead of relying on single sequences
carrying isolated pieces of information, we investigate ways in which
information can be shared, reinforced, and recovered through patterns of
relationships between many molecules. By taking inspiration from networked
systems, we aim to develop multiple complementary strategies that improve
robustness, scalability, and readout efficiency while remaining compatible
with existing molecular tools. These ideas open the door to dna storage
architectures that circumvent the limitations of classical direct storage with
DNA synthesis.